PhD Top Stories
Elena genovese
Biomarkers and precision therapy for primary immunodeficiencies: an in vitro study based on Induced Pluripotent Stem Cells from patients
Doctoral Programme in Reproduction and Developmental Sciences
In 2006 Takahashi and Yamanaka introduced to the scientific world a new type of stem cells, the so-called induced pluripotent stem cells or iPSCs (Nobel prize for medicine in 2012). These cells can be easily obtained by exposing in vitro to a series of genetic stimuli, a process called “reprogramming”, somatic cells including peripheral blood mononucleate cells that can be isolated from just few milliliters of patients’ blood, avoiding invasive procedures. The 4 reprogramming factors used as genetic stimuli are able to force somatic cells to an embryonic-like state obtaining cells able to differentiate into all 3 germ layers and into any adult cell type like embryonic stem cells. iPSCs retaining the genetic background of the patient and with their characteristic ability to differentiate under adequate stimuli into any cell type, represent a great tool for creating patient-specific models. For more than a decade, researchers have been using patient-specific iPSCs to model diseases caused by genetic defects. However, studies evaluating the pharmacological effects in this setting are still limited. In particular, by using iPSCs, it is possible to investigate the mechanisms and cytotoxicity of drugs potentially active in rare diseases. Rare diseases are often genetically based and developing therapies can be particularly challenging given the limited number of patients (1 per 2000). iPSCs can be a groundbreaking source of cells and tissues representative of these rare conditions, harboring the genomic alterations of interest, to perform the experiments needed to develop innovative personalized therapies.
In this context we set up an in vitro model of Aicardi Goutières (AGS) and Ataxia Telangiectasia (AT) syndromes, two rare inherited disorders of immunity with prevalent neurological symptoms (https://www.agsamericas.org), using patient iPSCs and investigated the cytotoxicity of different potentially effective immunomodulators. iPSCs were obtained by reprogramming AT and 3 forms of AGS (AGS1, AGS2, AGS7) patients’ cells and, as a control, the BJ normal human fibroblast line. Cytotoxicity of dexamethasone and mepacrine, immunosuppressive drugs proposed for treating AT and AGS respectively, were analyzed. Data were obtained also for other immunomodulatory drugs (thioguanine, mercaptopurine, thalidomide, and lenalidomide). Relative expression of genes involved in the tested drug pathways was determined and compared with control cell lines (figure 1), chosen principally on their known in vitro sensitivity to the drugs considered. HPRT1 was evaluated as marker for thiopurines in vitro effects due to its importance in thiopurines activation, resulting 50% lower in the BJ-iPSC line in comparison with the CCRF-CEM sensitive line. NR3C1, the gene coding for the glucocorticoid intracellular receptor for dexamethasone, was around 97% lower with respect to the NALM6 sensitive line; MB21D1 coding for cGAS, a crucial enzyme in the pathway leading from the sensing of cytoplasmic nucleic acids to the production of interferons, resulted around 50% lower in the BJ-iPSC in comparison to the sensitive monocytic leukemia THP1 line and significantly higher in the AGS7 compared to BJ-iPSC. As for thalidomide and lenalidomide, a gene coding for a ubiquitin ligase, cereblon (CRBN), was analyzed, being identified as the principal target of their pharmacological effects, resulting 350% higher in BJ-iPSC than in the THP1 cells.
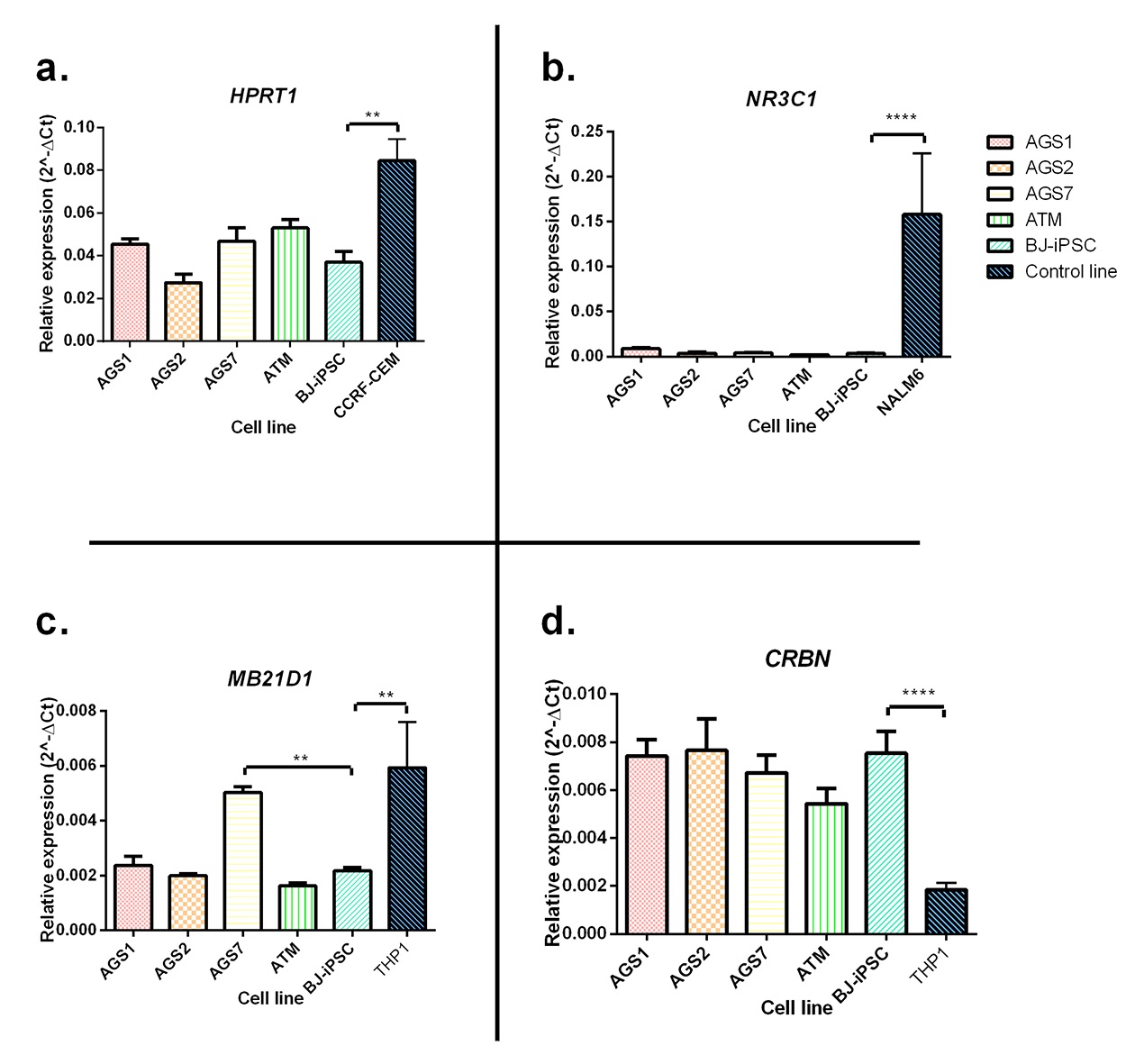
Figure 1: Real-time PCR (polymerase chain reaction) analysis of genes involved in tested drug pathways. (a) HPRT1: key gene in thiopurine drugs activation. (b) NR3C1: gene coding for glucocorticoid intracellular receptor. (c) MB21D1: gene coding for cGAS, important enzyme in mepacrine activity. (d) CRBN: gene involved in thalidomide and lenalidomide pharmacological effects. The data are represented as relative expression with respect to the housekeeping gene ACTB (2-?Ct) and are reported as means ± SE of three independent experiments. Statistical difference: **, P < 0.01; ****, P < 0.0001 BJ-iPSC line vs. all other lines (one-way ANOVA (analysis of variance) and Bonferroni’s posttest comparing all lines with BJ-iPSC as control).
Overall, no correlation between relative expression of genes analyzed and cytotoxicity was identified except for MB21D1 in the AGS7 line. Indeed, AGS7 iPSCs displayed altered viability (figure 2) when treated with a low dose of mepacrine and higher expression of MB21D1, which is the main target for mepacrine action. AGS7 iPSCs were also more sensitive to thioguanine, while AGS2 and AT iPSCs were less sensitive to this medication than the BJ-iPSC (figure 2). All iPSCs were equally extremely sensitive to mercaptopurine and resistant to dexamethasone, thalidomide, and lenalidomide.
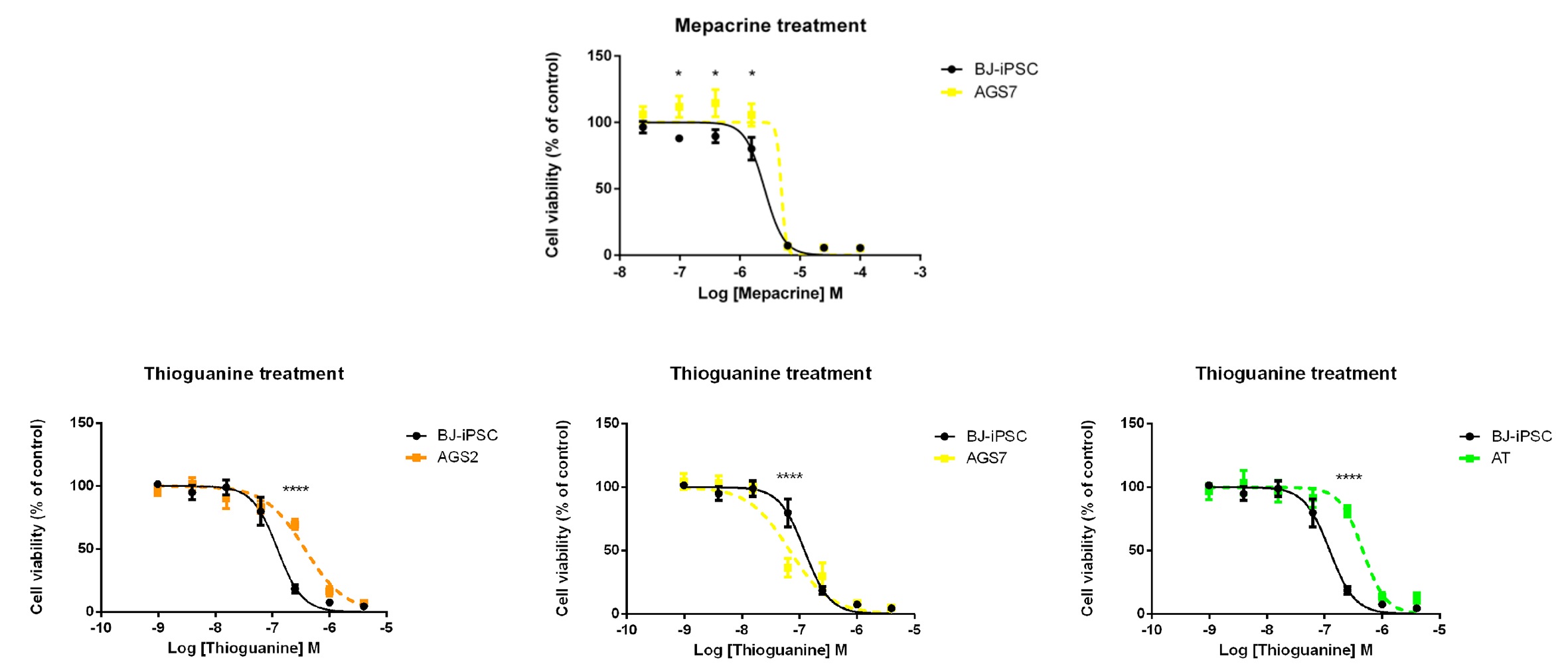
Figure 2: Cytotoxic effect of mepacrine AGS7, and thioguanine on AGS2, AGS7 and AT patient-derived iPSCs (color) in comparison with the BJiPSC control line (black). Cells were exposed for 72 hours to drugs, and cytotoxic effects were analyzed by MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. The data are reported as percentage of cell viability with respect to untreated cells (cells not exposed to the drug) and are the means ± SE of four independent experiments performed in triplicate. Statistical analysis vs. BJ-iPSC control line: *, P < 0.05 (two-way ANOVA (analysis of variance), and Bonferroni’s posttest).
With this work we set up an in vitro model of patient-specific iPSCs useful to investigate the mechanisms of drugs potentially effective in AT and AGS. Considering that the number of studies on this topic is still limited, our work lays the foundation for future insight on patient-specific therapy and rare diseases to extend the study to tissues involved in AT and AGS pathogenesis, such as the immune or nervous system cells by differentiating iPSCs.
Authors and affiliations
Elena Genova1, Federica Cavion1, Marianna Lucafò2, Marco Pelin1, Gaetana Lanzi3,4, Stefania Masneri3,4, Rosalba Monica Ferraro3,4, Elisa Maria Fazzi5,6, Simona Orcesi7,8, Giuliana Decorti2,9, Alberto Tommasini2, Silvia Giliani3,4, Gabriele Stocco12Institute for Maternal and Child Health IRCCS Burlo Garofolo, Trieste, Italy
3“Angelo Nocivelli” Institute for Molecular Medicine, ASST Spedali Civili, Brescia, Italy
4Department of Molecular and Translational Medicine, University of Brescia, Brescia, Italy
5Child Neurology and Psychiatry Unit, ASST Spedali Civili, Brescia, Italy
6Department of Clinical and Experimental Sciences, University of Brescia, Brescia, Italy
7Department of Brain and Behavioral Sciences, University of Pavia, Italy
8Child Neurology and Psychiatry Unit, IRCCS Mondino Foundation, Pavia, Italy
9Department of Medical, Surgical and Health Sciences, University of Trieste, Trieste, Italy.
Contact
Elena Genova, email: elena.genova@phd.units.itReference
Elena Genova, Federica Cavion, Marianna Lucafò, Marco Pelin, Gaetana Lanzi, Stefania Masneri, Rosalba Monica Ferraro, Elisa Maria Fazzi, Simona Orcesi, Giuliana Decorti, Alberto Tommasini, Silvia Giliani and Gabriele StoccoBiomarkers and Precision Therapy for Primary Immunodeficiencies: An In Vitro Study Based on Induced Pluripotent Stem Cells From Patients
Clinical Pharmacology & Therapeutics 108, 358-367 (2020)
DOI: 10.1002/cpt.1837
Informazioni aggiornate al: 28.9.2020 alle ore 11:38
Contact: Webmaster - Università di Trieste pagina curata da: Research Doctorate

Piazzale Europa, 1 - 34127 - Trieste, Italia -
Tel. +39 040 558 7111 - P.IVA 00211830328
C.F. 80013890324 - P.E.C. ateneo@pec.units.it



