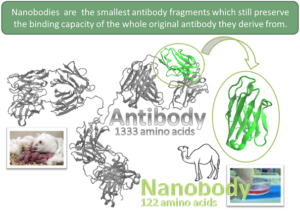
Nanobody design
We computationally optimize small antibody fragments (VHH or nanobodies) capable of recognizing chosen epitopes with the aim of tailoring novel binders for medical and pharmaceutical applications. Evolved VHHs are then produced and analyzed for their affinity and stability in an iterative two-way transfer between empiric data and theoretical modeling [a,b]. We further study their humanization with the final aim of developing new anticancer drugs. Up to date our main achievement is the development of a computational protocol capable generating VHHs with nanomolar binding affinity towards a chosen epitope of a protein target [c]. We carry out this activity thanks to computational projects granted by CINECA (the Italian supercomputing centre for scientific research) and PRACE (Partnership for Advanced Computing in Europe), and funding from the European Social Fund, the Regional Operative Programme, Alternatives Research & Development Foundation (ARDF), and the Italian Association for Cancer Research (AIRC).
Current collaborators: |
Molecular Modeling for Medicinal ChemistryWe support medical doctors (and a collaborating network that includes the group of J.E. Rothman, 2013 Nobel Prize in Medicine) in modeling protein mutants [a,b], and synthetic medicinal chemists in modeling potential drugs [c] for a variety of systems of medical interest.
Current collaborators: |
New architectures for biosensing
Current collaborators: |
Molecular and colloidal self-assembly
Current collaborators: |
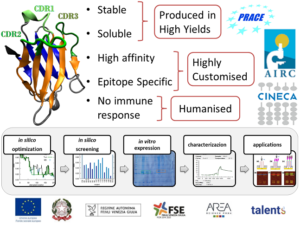
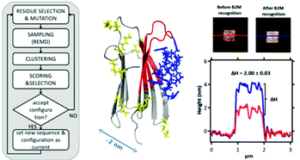 The design of new strong and selective binders is a key step towards the development of new sensing devices and effective drugs. Enhanced affinity and selectivity can be achieved through careful design [b,d]. In this framework I am coordinating the joint theoretical/experimental development of novel peptide-based computationally designed nanodevces for protein recognition. The group is developing new protocols based on Monte Carlo, docking, and molecular dynamics for the design and screening of peptides for protein recognition [a,c]. Up to date our main achievement is the development of a computational protocol capable of generating peptides for the selective detection of a chosen epitope of a protein target [a,c]. This activity started in the framework of the 7FP, Ideas, ERC Advanced Grant: Molecular Nanotechnology For Life Science Application: Quantitative Interactomics for Diagnostics, Proteomics and Quantitative Oncology, (Quidroquo) (proposal n. 269025, 2011-2016 PI: G. Scoles), and continued through a number of computational projects granted by CINECA, the Italian supercomputing centre for scientific research, and funding from the Italian Association of Cancer Research, and the Royal Society of Chemistry (UK).
The design of new strong and selective binders is a key step towards the development of new sensing devices and effective drugs. Enhanced affinity and selectivity can be achieved through careful design [b,d]. In this framework I am coordinating the joint theoretical/experimental development of novel peptide-based computationally designed nanodevces for protein recognition. The group is developing new protocols based on Monte Carlo, docking, and molecular dynamics for the design and screening of peptides for protein recognition [a,c]. Up to date our main achievement is the development of a computational protocol capable of generating peptides for the selective detection of a chosen epitope of a protein target [a,c]. This activity started in the framework of the 7FP, Ideas, ERC Advanced Grant: Molecular Nanotechnology For Life Science Application: Quantitative Interactomics for Diagnostics, Proteomics and Quantitative Oncology, (Quidroquo) (proposal n. 269025, 2011-2016 PI: G. Scoles), and continued through a number of computational projects granted by CINECA, the Italian supercomputing centre for scientific research, and funding from the Italian Association of Cancer Research, and the Royal Society of Chemistry (UK).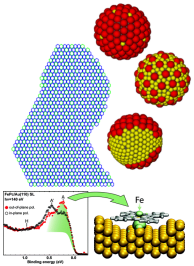 Molecules do self-organize into ordered molecular structures thanks to the intermolecular forces acting between them. We look at their organization at different resolutions through a variety of computational approaches: coarse grained models for the simplified representations of large molecular ensembles [a], atomistic resolution models [b] , and quantum mechanical methods to study the effect of the electrons on the molecules under investigation [c,d]. Our latest achievement is the mapping of the self-assembled structures that can emerge when molecules can access multiple adsorption states [e].
Molecules do self-organize into ordered molecular structures thanks to the intermolecular forces acting between them. We look at their organization at different resolutions through a variety of computational approaches: coarse grained models for the simplified representations of large molecular ensembles [a], atomistic resolution models [b] , and quantum mechanical methods to study the effect of the electrons on the molecules under investigation [c,d]. Our latest achievement is the mapping of the self-assembled structures that can emerge when molecules can access multiple adsorption states [e].